How bacteria exploit a chink in the body's armor
Advertisement
Scientists have discovered how a unique bacterial enzyme can blunt the body's key weapons in its fight against infection.
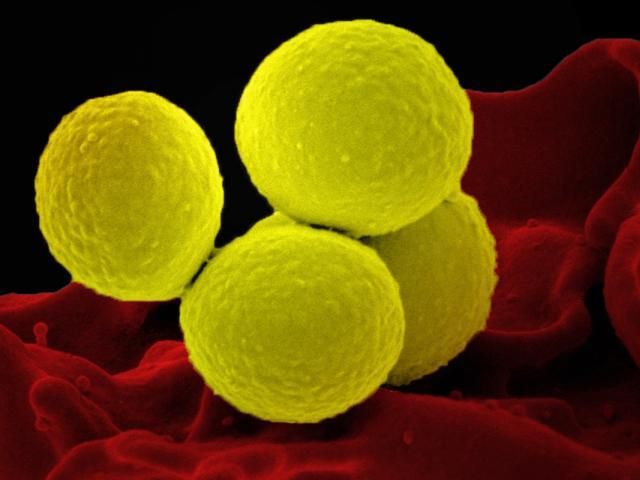
Staphylococcus aureus, in yellow, interacts with a human white blood cell.
Photo courtesy the National Institute of Allergy and Infectious Disease
Researchers at the University of Illinois at Urbana-Champaign and Newcastle University in the U.K. are investigating how infectious microbes can survive attacks by the body's immune system. By better understanding the bacteria's defenses, new strategies can be developed to cure infections that are currently resistant to treatments, the researchers said.
The study focused on the bacterium Staphylococcus aureus, which is found on approximately half of the population. While it usually safely coexists with healthy individuals, S. aureus has the ability to infect nearly the entire body; in its most pathogenic form, the bacterium is the so-called "superbug" methicillin-resistant S. aureus, or MRSA.
The human body uses a diverse array of weapons to fight off bacteria like S. aureus. "Our immune system is very effective and prevents the majority of microbes we encounter from causing infections," said U. of I. microbiology professor Thomas Kehl-Fie , who led the study with Kevin Waldron, of Newcastle University. "But pathogens such as S. aureus have developed ways to subvert the immune response."
S. aureus can overcome one of the body's key defenses, nutritional immunity, which prevents bacteria from obtaining critical nutrients. It starves S. aureus of manganese, a metal needed by the bacterial enzyme superoxide dismutase, or SOD. This enzyme functions as a shield, minimizing the damage from another weapon in the body's arsenal, the oxidative burst. Together, the two host weapons usually function as a one-two punch, with nutritional immunity weakening the bacteria's shields, enabling the oxidative burst to kill the bacterium.
S. aureus is particularly adept at causing devastating infections. Differing from other closely related species, S. aureus possesses two SOD enzymes. The team discovered that the second SOD enhances the ability of S. aureus to resist nutritional immunity and cause disease.
"This realization was both exciting and perplexing, as both SODs were thought to utilize manganese and therefore should be inactivated by manganese starvation," Kehl-Fie said.
The most prevalent family of SODs, to which both of the S. aureus enzymes belong, has long been thought to come in two varieties: those that are dependent on manganese for function and those that use iron.
In light of their findings, the team tested whether the second staphylococcal SOD was dependent on iron. To their surprise, they discovered that the enzyme was able to use either metal. While the existence of these cambialistic SODs, capable of using both iron and manganese, was proposed decades ago, the existence of this type of enzyme was largely dismissed as a quirk of chemistry, unimportant in real biological systems. The team's findings dispel this notion, demonstrating that cambialistic SODs critically contribute to infection.
The team found that, when starved of manganese by the body, S. aureus activated the cambialistic SOD with iron instead of manganese, ensuring its critical bacterial defensive barrier was maintained.
"The cambialistic SOD plays a key role in this bacterium's ability to evade the immune defense," Waldron said. "Importantly, we suspect similar enzymes may be present in other pathogenic bacteria. Therefore, it could be possible to target this system with drugs for future antibacterial therapies."
The emergence and spread of antibiotic-resistant bacteria, such as MRSA, make such infections increasingly difficult, if not impossible, to treat.
This has prompted leading health organizations, such as the Centers for Disease Control and Prevention and the World Health Organization, to issue an urgent call for new approaches to combat the threat of antibiotic resistance.