Viruses glue RNA to proteins
Advertisement
Until now, RNA and proteins were thought to interact only briefly during cellular processes. Researchers at the Max Planck Institute for Terrestrial Microbiology in Marburg, Germany, have discovered that this is not the case. During their developmental cycle, bacterial viruses `glue` specific RNAs to host proteins. As the authors descibe in their publication in the journal "Nature",`RNAylation` could open up new avenues for phage therapy or drug development.
"Life is a relationship between molecules," wrote the famous biologist Linus Pauling. Interactions between proteins and RNA (ribonucleic acids) affect translation, repair of genetic information, and transport of cellular building blocks. These interactions are transient contacts between RNA and RNA-binding proteins based on specific RNA structures or sequences.
Now, a team of researchers at the Max Planck Institute in Marburg, Germany, has discovered that protein and RNA can also be tightly bound to each other in a so-called covalent bond.
Bacteriophages as “fast killers”
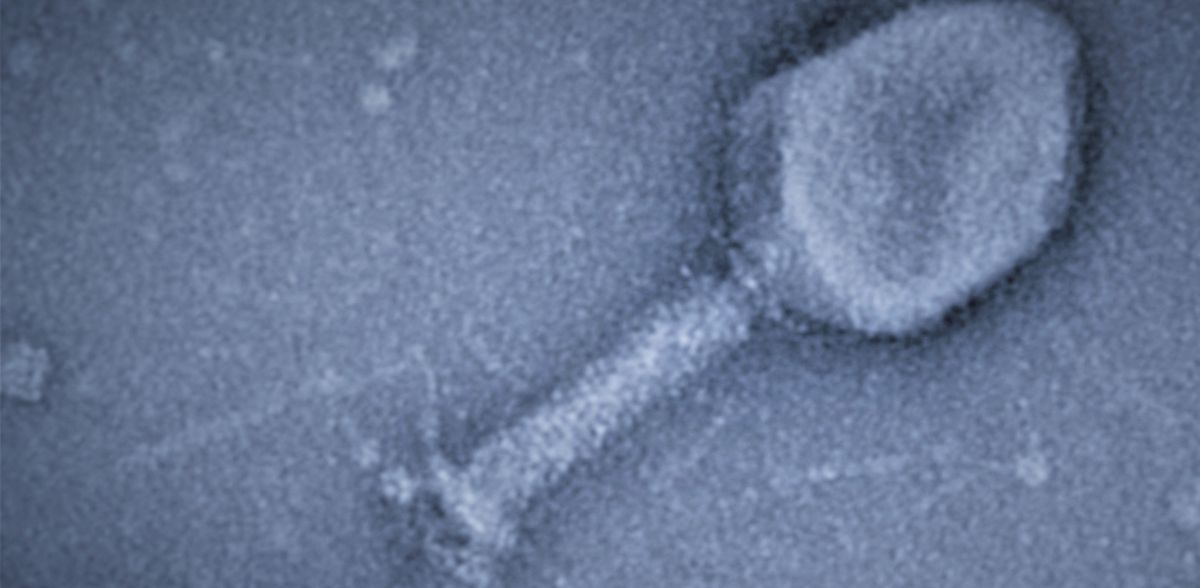
In their study, published in the current issue of the journal Nature, the research group led by Dr. Katharina Höfer examined a system of bacteria and bacterial viruses (bacteriophages). The latter attack very specific bacteria, such as the T4 phage that infects the bacterium E. coli. T4 is a "fast killer": the bacterial cell is destroyed 20 to 30 minutes after infection begins. This is faster than an antibiotic works. With antibiotic resistance on the rise, phage therapy is being explored as a potential alternative for treating bacterial infections.
To infect the bacterium, bacteriophage T4 has evolved fascinating strategies. After invasion, it uses three different ADP-ribosyltransferases (ARTs) as biocatalysts. By attaching a part of the coenzyme nicotinamide adenine dinucleotide (NAD) to proteins, these ARTs modify more than 30 host proteins. This enables the phage to reprogram and kill the bacterium.
NAD-RNA connects RNA- and phage research
Katharina Höfer has been studying the function of RNA for some time. She is particularly interested in NAD-RNAs, which are RNAs that carry an NAD attachment. Eight years ago, she and her colleagues at the University of Heidelberg discovered that this type of RNA occurs in bacteria. Since then, NAD-capped RNAs have been found in many different forms and sizes and in different groups of organisms, but their biological significance remained unclear.
Katharina Höfer wondered whether an ADP-ribosyltransferase such as that used by the T4 phage could attach not only NAD but also NAD-RNA to proteins. To answer this question, the researchers had to develop many methods themselves. But then it became clear: The ART ModB of the T4 phage accepts not only NAD, but also NAD-RNA as a substrate - both in the test tube and in vivo, in the living system. The researchers called this novel reaction - the binding of a whole RNA to a protein - RNAylation. It is a completely new concept of natural RNA-protein interaction.
RNAylation may be a mechanism for control of cellular resources
But why does the T4 phage use RNAylation? Apparently, this process is essential for efficient phage infection, because mutants of the T4 phage that lack ModB kill bacteria much more slowly. The research group was able to show that in living cells, ModB specifically binds different RNAs to bacterial proteins involved in translation. Maik Wolfram-Schauerte, first author of the study, suggests: "RNAylation may be part of the phage's strategy. The attachment of bacterial RNA to ribosomes may stop the translation of bacterial proteins, which enables the phage to regulate the biosynthesis of its own proteins."
RNAylation as a potential new tool for synthetic biology
To investigate the molecular mechanisms of RNAylation, Katharina Höfer started a collaboration with researchers at the University of Heidelberg and the Max Planck Institute for Multidisciplinary Sciences in Göttingen.
Katharina Höfer explains: "Our results not only extend the previous picture of the developmental cycle of phages. They point to a completely new biological role for NAD-modified RNA, namely the activation of RNA for enzymatic transfer to a protein. This also opens up new avenues of research.”
For example, RNAylation could become a tool for synthetic biology in the future. As a "molecular glue," it could be used to form specific RNA-protein conjugates to exploit the properties of proteins and nucleic acids in combination.
But, there are still many unanswered questions. "Some ARTs accept NAD-RNA, others do not - this raises the question of the exact mechanism," explains Katharina Höfer. "The difficulty is that the modification is quite large and complex. In the test tube, RNAylation is relatively easy to detect, but in vivo, the diversity of target proteins and RNAs makes it challenging to study. To elucidate the function of RNAylation, we need to develop new methods to study our specific questions in the living system.”