How the gut microbiome responds to antibiotics
Advertisement
antibiotics affect the composition and dynamics of the gut microbiome. Treatment with antibiotics not only leads to a loss of biodiversity of microorganisms, but also often favours the selection of resistant strains of bacteria. It has been unclear how the microbiome responds to repeated antibiotic therapy. In a preclinical study, an international research team led by two DZIF scientists used metagenome and cultivation analyses to identify evolutionary mechanisms that contribute to the resilience of the microbial community after repeated antibiotic administration. The study has now been published in the renowned scientific journal Cell Host & Microbe.
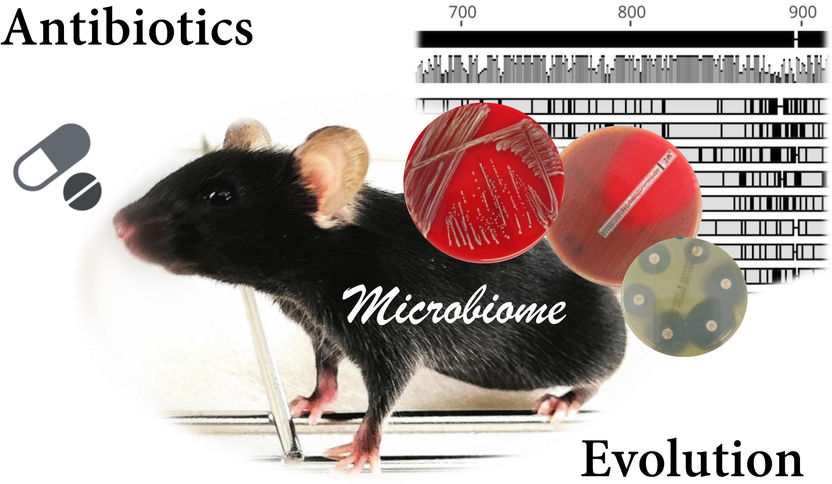
Illustration of the described microbiome research: germ-free mouse as an animal model, agar plates with cultures of bacteria isolated from the intestine and antibiotic test strips and platelets, and the representation of a DNA sequence comparison.
Bärbel Stecher, Max von Pettenkofer Institute, LMU Munich
Each person's gut microbiome contains a specific community of microorganisms that normally remains stable for years. However, it can be thrown off balance by factors such as dietary changes, infections or medications. Antibiotics in particular have a strong influence on the microbiome. In response, microorganisms employ various resistance mechanisms, with individual bacterial populations evolving through selection of antibiotic-resistant variants. Yet, the extent and mechanisms of these processes and their impact on the ecology of the microbial community are poorly understood.
In a comprehensive metagenomic study, DZIF scientists Prof. Bärbel Stecher and Prof. Alice McHardy, together with an international research team, investigated the evolution of intestinal bacteria exposed to repeated disruptions by antibiotics. For this purpose, they used a gnotobiotic mouse model, i.e., mice kept germ-free and stably colonised with a known consortium of bacteria. This model allows evolutionary studies of individual members of the community in the natural host under well-defined and controllable conditions. The researchers then analysed the effects of different classes of antibiotics on the microbiome over a period of 80 days. Using metagenomic analyses, they followed the selection of putative antibiotic resistance-promoting mutations in the bacterial populations, and subsequently analysed the characteristics of evolved bacterial clones isolated from the communities.
"We were able to track how repeated antibiotic therapy leads to the selection of antibiotic-resistant commensal bacteria, which after a while increases the resilience of the microbial community to certain antibiotics such as the tetracyclines. In addition to adaptation of the microbiome through evolution of individual microorganisms, we also found evidence of resistance development of individual bacteria through slowing of cell growth. The microbiome adapts to the treatment, so to speak, and is better able to withstand it," says Bärbel Stecher, coordinator of the Gastrointestinal Infections research area at the German Center for Infection Research (DZIF) and professor of Medical Microbiology and Hygiene at the Max von Pettenkofer Institute at Ludwig-Maximilians-Universität München (LMU).
In addition, the research team observed an induction of prophages triggered by treatment with certain antibiotics. In this process, lysogenic bacteriophages—whose genomes are integrated into bacterial genomes—are activated, whereupon they proliferate and lyse the host cells upon release of new viral particles. "This is an example of how antibiotics can also indirectly affect bacterial survival," says Dr Philipp Münch, first author of the study.
Overall, the study shows an immense diversity in the response of the microbiome to antibiotic treatments. This includes, for example, ecological effects such as the inhibition of a microorganism by the elimination of an important "partner" bacterium in the metabolic network of the gut ecosystem.
"Due to this high complexity of direct and indirect responses, it is difficult to predict which species will be affected by treatment with an antibiotic, even in gnotobiotic animal models with a defined community of microorganisms," summarises Prof. Alice McHardy, Deputy Coordinator Bioinformatics and Machine Learning at DZIF and head of the Department of Computational Biology for Infection Research at the Helmholtz Centre for Infection Research, a member institution of the DZIF.