Study pinpoints SARS-CoV-2 Spike mutation that ‘escapes’ killer T-cells generated by infection and vaccination
Cardiff University scientists urge monitoring of ‘viral escape’ – and warn vaccines may need altering
Advertisement
UK scientists have highlighted a mutation in the spike protein of SARS-CoV-2 that allows the virus to evade important immune cells induced by infection and vaccines.
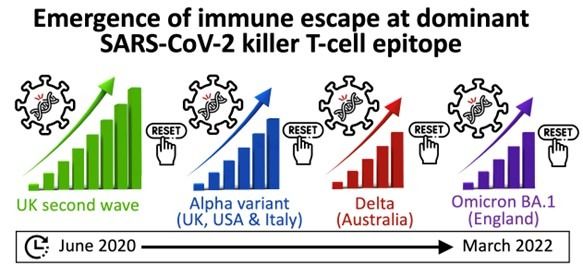
The P272L Spike mutation first arose during the UK’s second wave of COVID-19, which began in September 2020, and has been pinpointed to the part of the Spike protein most frequently recognised by killer T-cells. The SARS Spike protein is also the basis for current vaccines.
The “escape mutation” has been found in more than 100 viral lineages to date, including those classed as “variants of concern”. It has been seen in strains in the UK and Europe, and in Australia and the USA between September 2020 and March 2022.
When a mutation arises independently multiple times this raises concerns the virus is evolving for a reason such as immune escape.
Now, a new study led by Cardiff University and published in Cell, has shown that the P272L Spike mutation escaped recognition by killer T-cells in a group of healthcare workers from south Wales infected with SARS-CoV-2 early in the pandemic. The mutation also evaded all T-cells raised against this part of the virus in donors that had been vaccinated.
The researchers say monitoring for “viral escape” is important – and if mutations like P272L start to dominate then future vaccines may need to be altered to include different viral proteins.
Lead author Professor Andrew Sewell, from Cardiff University’s School of Medicine and Systems Immunity Research Institute, said: “We studied more than 175 different types of killer T-cells that could see the part of the virus containing the P272L mutation and were surprised to see that this one mutation resulted in escape in all the donors studied.
“Similar escape mutations have been seen in influenza viruses. It has been estimated the virus that caused the 1968 Hong Kong flu pandemic has mutated to escape from recognition by a type of killer T-cell once every three years.”
In this study, P272L escaped from more than 175 T-cell receptors – both in the COVID patients and those who had been vaccinated.
Since the Cardiff study was completed, the mutation has been observed in the original Omicron variant (BA.1) in England. This variant was outcompeted by other Omicron variants (most recently BA.5) that are more transmissible, but the researchers anticipate that the killer T-cell escape mutant may arise and transmit in BA.5 and any subsequent variants.
Professor Tom Connor, an expert in SARS-CoV-2 genomics from Cardiff University’s School of Biosciences and Systems Immunity Research Institute, said: “The P272L mutation has arisen in many different SARS-CoV-2 lineages since the start of the pandemic. The independent evolution of the same mutation in multiple viral lineages is an example of homoplasy, which may be suggestive of a mutation that could confer a selective advantage.
“I anticipate that we may see more occurrences of the P272L variant over time, as the virus continues its evolution into a human pathogen. Our work emphasises how much we still have to learn about SARS-CoV-2, and the importance of monitoring T-cell escape in future.”
Dr Lucy Jones, clinical research lead from Cwm Taf Morgannwg University Health Board who collected the study samples, said: “Our study looked at recognition of the virus spike protein by the most common sort of killer T-cell across the population. If the P272L mutation comes to predominate in time, it might be advantageous to provide vaccine boosts with this new sequence or to extend vaccination to include other viral proteins.
“Current vaccines work well and should still be taken up by people, but our research suggests vaccines may need altering as we learn more about the virus.”