Predicting a protein's behavior from its appearance
Advertisement
proteins are the building blocks of life and play a key role in all biological processes. Understanding how they interact with their environment is therefore vital to developing effective therapeutics and the foundation for designing artificial cells.
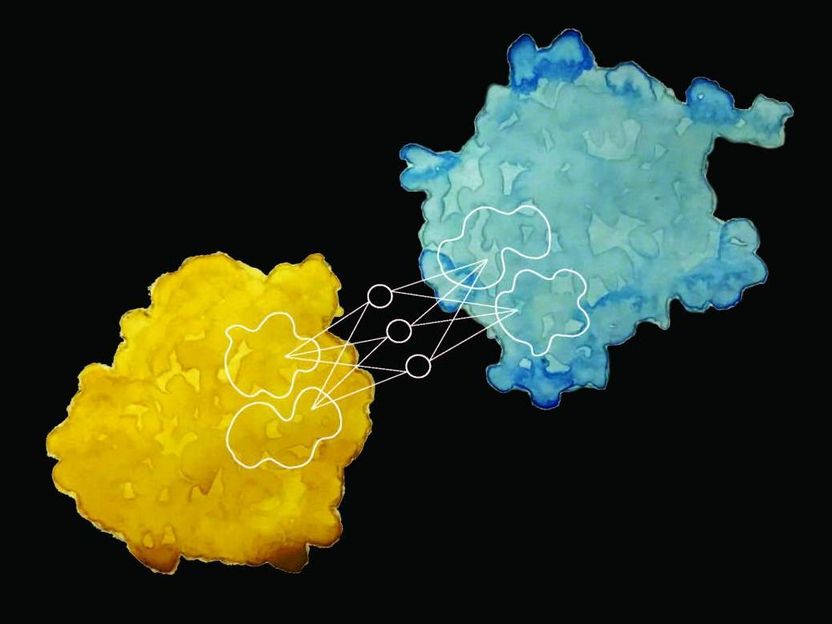
Researchers at EPFL have developed a new way to predict a protein's interactions with other proteins and biomolecules, and its biochemical activity, merely by observing its surface.
Laura Persat / 2019 EPFL
Researchers at the Laboratory of Protein Design & Immunoengineering (LPDI), part of EPFL's Institute of Bioengineering at the School of Engineering, working with collaborators at USI-Lugano, Imperial College and, Twitter's Graph Learning Research division have developed a groundbreaking machine learning-driven technique for predicting these interactions and describing a protein's biochemical activity based on surface appearance alone. In addition to deepening our understanding of how proteins function, the method - known as MaSIF - could also support the development of protein-based components for tomorrow's artificial cells.
Data-driven research
The researchers took a vast set of protein surface data and fed the chemical and geometric properties into a machine-learning algorithm, training it to match these properties with particular behavior patterns and biochemical activity. They then used the remaining data to test the algorithm. "By scanning the surface of a protein, our method can define a fingerprint, which can then be compared across proteins," says Pablo Gainza, the first author of the study.
The team found that proteins performing similar interactions share common "fingerprints."
"The algorithm can analyze billions of protein surfaces per second," says LPDI director Bruno Correia. "Our research has significant implications for artificial protein design, allowing us to program a protein to behave a certain way merely by altering its surface chemical and geometric properties."
The method, published in open-source format, could also be used to analyze the surface structure of other types of molecules.
Original publication
P. Gainza, F. Sverrisson, F. Monti, E. Rodolà, D. Boscaini, M. M. Bronstein, and B. E. Correia; "Deciphering interaction fingerprints from protein molecular surfaces using geometric deep learning"; Nature Methods; 2019.
Other news from the department science
Most read news
More news from our other portals
See the theme worlds for related content
Topic world Protein analytics
Protein analytics provides a deep insight into these complex macromolecules, their structure, function and interactions. It is essential for discovering and developing biopharmaceuticals, understanding disease mechanisms, and identifying therapeutic targets. Techniques such as mass spectrometry, Western blot and immunoassays allow researchers to characterize proteins at the molecular level, determine their concentration and identify possible modifications.

Topic world Protein analytics
Protein analytics provides a deep insight into these complex macromolecules, their structure, function and interactions. It is essential for discovering and developing biopharmaceuticals, understanding disease mechanisms, and identifying therapeutic targets. Techniques such as mass spectrometry, Western blot and immunoassays allow researchers to characterize proteins at the molecular level, determine their concentration and identify possible modifications.