Approved medications – new role in combating infections?
Advertisement
influenza A viruses periodically cause severe influenza epidemics. Due to their genetic variability, new subtypes can emerge that are no longer covered by seasonal influenza vaccines. Looking for an effective treatment of such serious influenza infections, researchers of the Paul-Ehrlich-Institut have used bioinformatics to screen licensed compounds for their potential efficacy against influenza viruses. Promising candidates were then tested in cell-based and animal studies, and dextromethorphan showed the most potential for further development.
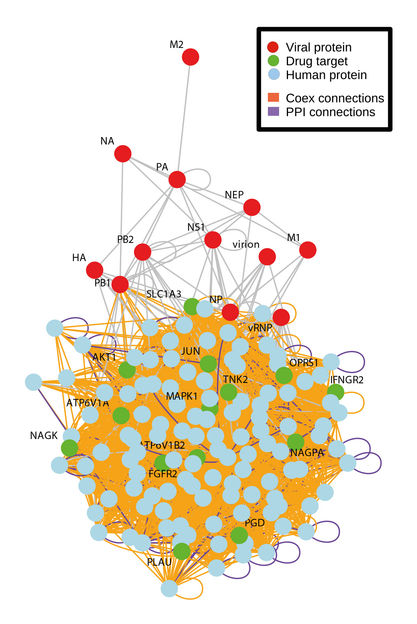
Using bioinformatics in search of effective medicinal products against influenza.
PEI
Influenza A virus ranks among the most feared pathogens. It regularly causes influenza epidemics. Its envelope contains the ion-channel forming M2 protein as well as the surface glycoproteins haemagglutinin (HA) and neurminidase (NA). Based on the antigenic properties of these glycoproteins, the viruses are categorised into different subtypes. Currently, 18 different haemagglutinin and 11 neuramidase subtypes have been identified. Influenza vaccines contain antigen components of the different circulating subtypes. However, the high genetic variability of influenza viruses requires annual vaccine updates.
If a new influenza virus emerges, the development and production of a matching vaccine will take several months. During this time, compounds that block the virus (antivirals) are the only treatment option. However, resistance against these compounds – M2 protein ion channel blockers and neuraminidase inhibitors – rapidly emerges, reducing their efficacy.
Repurposing of compounds originally licensed for other indications to treat diseases like influenza is a promising new approach. Unlike compounds that are designed to target the pathogen directly, this approach looks for compounds that target proteins in the infected cell that are needed for the replication of the virus. On the one hand, targeting cellular proteins is expected to be associated with less resistance development, and on the other hand, such a compound may be efficacious against a broad spectrum of virus strains.
So-called genome-wide screenings with “small interfering” RNAs (siRNAs) were used to identify candidate proteins and signaling pathways involved in the influenza virus replication cycle. In co-operation with researchers from various different research institutes in Singapore, and with support from the German Centre for Infection Research (DZIF), Professor Veronika von Messling, head of Division Veterinary Medicine of the Paul-Ehrlich-Institut until September 2018, and her group have compared these candidates with known targets of already licensed compounds, which would be immediately available in the event of an epidemic. The most promising candidates were then first tested in vitro (in cell culture) and followed by animal experiments with mice and ferrets in vivo.
Out of 15 candidate compounds, four were able to control an influenza A infection in vitro. In efficacy studies in mice, dextromethorphan led to a significant decrease in viral load in the lung and increased efficacy of the antiviral agent Oseltamivir. In ferrets infected with a seasonal H1N1 strain, the administration of dextromethorphan also reduced disease severity but not the virus titer in the lung.
“Our data show that dextromethorphan could be a therapeutic option for influenza”, said Professor von Messling. Furthermore, the study shows the potential of bioinformatics-based approaches to identify promising candidates for drug repurposing to treat infectious diseases.