Nucleosome breathing from atomistic time snapshots
Computer simulations visualize in atomic detail how DNA opens while wrapped around proteins
Advertisement
Researchers from the Hubrecht Institute in Utrecht (The Netherlands) and the Max Planck Institute for Molecular Biomedicine in Münster (Germany) used computer simulations to reveal in atomic detail how a short piece of DNA opens while it is tightly wrapped around the proteins that package our genome. These simulations provide unprecedented insights into the mechanisms that regulate gene expression.

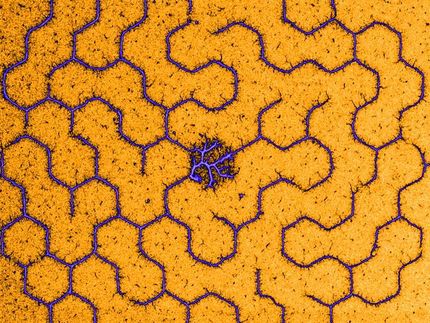
Three microseconds from the life time of a nucleosome: Snapshots in time were taken every 4 nanoseconds and were superimposed on the core region of the histones.
© Jan Huertas und Vlad Cojocaru; MPI Münster / Hubrecht Institute
Every cell in the body contains two meters of DNA. In order to fit all the DNA in the cell’s small nucleus, the DNA is tightly packed in a structure known as chromatin. Chromatin is an array of identical smaller structures named nucleosomes. In a single nucleosome, DNA is wrapped around 8 proteins called histones. Chromatin is not uniformly compact across the genome. The tightness of the packaging is important in regulating which genes are expressed and therefore which proteins are produced by a cell.
Transitions from tightly to loosely packed DNA – from closed to open chromatin – are essential for cells to convert to another cell type. These cell conversions are hallmarks of development and disease, but are also often used in regenerative therapies. Understanding how such transitions occur may contribute to understanding diseases and optimizing therapeutical cell type conversions.
Computational nanoscope
One step in the opening of chromatin is the motion of DNA while wrapped in nucleosomes. Like all molecular structures in our cells, nucleosomes are dynamic. They move, twist, breathe, unwrap and wrap again. Visualizing these motions using experimental methods is often very challenging. One alternative is to use the so-called “computational nanoscope”.
Researchers use the term computational nanoscope to refer to a set of computer simulation methods. These methods enable them to visualize the movements of molecules over time. Over the past years, the methods have become so accurate that researchers started referring to them as a computational nanoscope; observing the molecules moving on the computer is similar to observing them under a very high resolution nanoscope.
Nucleosomes breathing
Jan Huertas and Vlad Cojocaru, supported by Hans Schöler from the Max Planck Institute for Molecular Biomedicine (Münster, Germany), generated multiple real-time movies of the movements of nucleosomes, each covering one microsecond from the nucleosome lifetime. Using these movies, they monitored how the nucleosomes open and close in a motion known as nucleosome breathing.
In their new paper, published in PLoS Computational Biology, Huertas and Cojocaru describe what causes nucleosome breathing. First, they found that the order in which the building blocks of DNA are arranged – the DNA sequence – is important for nucleosome breathing. Second, the dynamics of histone tails are essential for this process. These histone tails are flexible regions in the histones that play a role in the regulation of gene expression. While the role of histone tails has been studied intensively, little is known about how they influence the motions of single nucleosomes. With their simulations, Huertas and Cojocaru described the relationship between histone tails and nucleosome breathing in atomic detail.
Histone modifications
“Being able to observe the breathing of nucleosomes in computer simulations is very challenging. The fact that we have now been able to visualize this represents a major step towards simulating the complete spectrum of nucleosome dynamics, from breathing to unwrapping. It also allows us to study how these motions are affected by modifications of the histones, which occur in different cells and regions of our DNA. Our simulations revealed that two histone tails are responsible for keeping the nucleosome closed. Only when these flexible tails moved away from particular regions of DNA, the nucleosome was able to open,” says the research leader, Vlad Cojocaru.
Jan Huertas, first author in the publication and recent PhD-graduate, adds: “Active (open) and inactive (closed) chromatin contain different modifications of histone tails. The next step is to perform simulations with such modifications. The atomic resolution of the simulations would allow us to pinpoint how each modification affects nucleosomes and chromatin dynamics.”
Towards understanding epigenetics
All three researchers are excited about the future of the use of atomistic computer simulations in understanding gene expression mechanisms in development and disease. “With the further increase of computational power available in the world, we will soon be able to simulate milliseconds of a nucleosome lifetime with all its atoms included. Furthermore, we will be able to routinely simulate multiple nucleosomes to study the effect of different modifications of histones on gene expression. This will give unprecedented insights into the mechanisms that regulate gene expression,” Cojocaru concludes.