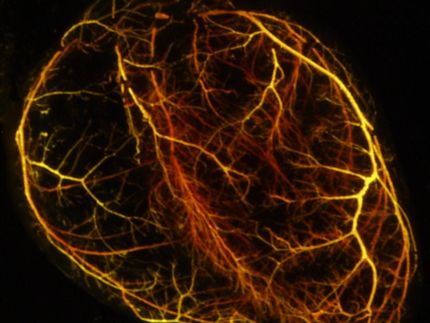
Cardioids – Heartbeat, heartbreak and recovery in a dish
Technology has led to the creation of spin-off HeartBeat.bio
Advertisement
Self-organizing heart organoids developed at IMBA – Institute of Molecular Biotechnology of the Austrian Academy of Sciences – are also effective injury- and in vitro congenital disease models. These “cardioids” may revolutionize research into cardiovascular disorders and malformations of the heart. The results are published in the journal Cell.
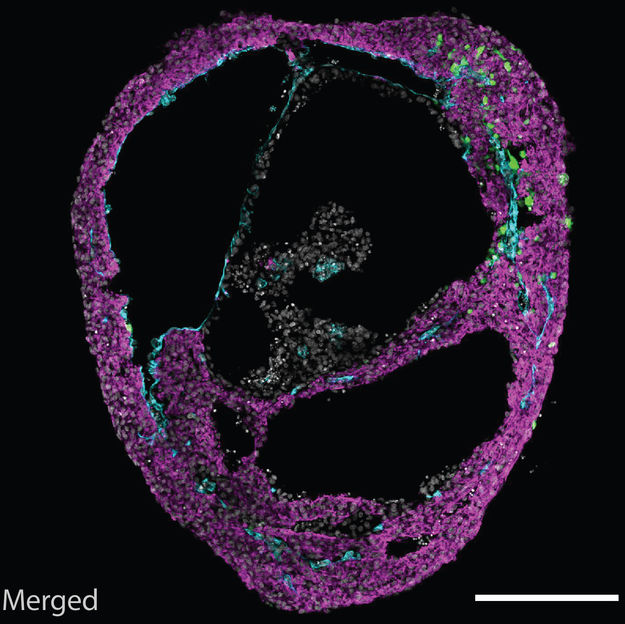
A self-organizing cardioid, containing all three cardiac lineages.
©Mendjan/IMBA
About 18 million people die each year from cardiovascular diseases, making them the leading cause of fatalities globally. Moreover, the most prevalent birth defects in children pertain to the heart. Currently, a major bottleneck in understanding human heart malformations and developing regenerative therapies are missing human physiological models of the heart.
The research group of Sasha Mendjan established cardioids at IMBA – human self-organizing cardiac organoids that recapitulate the lineage architecture of a heart chamber. Mendjan explains: “Cardioids are a major milestone. Our guiding principle is that for an in vitro tissue to be fully physiological, it also needs to undergo organogenesis. We were able to achieve this, using the developmental principles of self-organization – which makes it such an exciting discovery”.
During development, a heart chamber emerges from the mesoderm germ layer. The researchers thus established in vivo-like mesodermal signaling conditions guiding pluripotent stem cells. “Amazingly, this led to self-organization of a heart chamber-like structure that was beating. For the first time, we could observe something like this in a dish. It is a simple, robust and scalable model, and does not require addition of exogenous extracellular matrix like many other organoid models,” explains Sasha Mendjan. Besides a beating myocardial layer, a functional heart also contains an inner endothelial lining that later contributes to heart vasculature, and an outer epicardial layer that directs heart growth and regeneration. Cardioids recapitulate this three layered structure, shaping a heart chamber-like structure.
The scientists determined how signaling and transcription factors control cardioid chamber formation. For instance, the team was able to phenocopy the dramatic chamber cavity loss observed in children with Hypoplastic Left Heart Syndrome in cardioids by disrupting a transcription factor linked to this defect. The researchers also assessed the effects of cryoinjury (injury by freezing), a technique mimicking myocardial infarction, on cardioids. For the first time in a dish, the team found that this injury triggers an in vivo-like accumulation of extracellular matrix proteins in cardioids, an early hallmark of both regeneration and fibrotic heart disease.
The self-organizing organoid field has revolutionized biomedical research over the past decade. However, the heart was the last major inner organ missing such a physiological model capable of recapitulating developmental and injury response processes. “Cardioids bear incredible potential to unravel human congenital heart defects. As the system is physiological and scalable, this opens up huge possibilities for drug discovery and regenerative medicine,” says Sasha Mendjan.
This technology developed at IMBA has led to the creation of HeartBeat.bio, a new IMBA spin-off focused on the development of a high-throughput 3D screening platform for heart failure and cardiomyopathies.
Original publication
Other news from the department science
Most read news
More news from our other portals
Something is happening in the life science industry ...
This is what true pioneering spirit looks like: Plenty of innovative start-ups are bringing fresh ideas, lifeblood and entrepreneurial spirit to change tomorrow's world for the better. Immerse yourself in the world of these young companies and take the opportunity to get in touch with the founders.